SimPhospho: a user-friendly tool to enhance phosphopeptide analysis
Improving phosphoproteome identification through simulation of phosphopeptide tandem mass spectra
22 April 2018
The biochemical process of protein phosphorylation plays a vital role in the regulation of many cellular processes including cell cycle, growth, apoptosis and signal transduction pathways. Given that the exact molecular site of a phosphorylation event determines its particular switching activity, validation of phosphorylation sites is of great importance.

The leading technology to discover and confirm protein phosphorylation is tandem mass spectrometry. It has been successfully employed for two decades to identify sites of phosphorylation. However, unambiguous phosphosite assignment still is considered challenging. A further improvement of sophisticated phosphopeptide data analysis strongly depends on the ability to interpret more complex tandem mass spectrometry spectra.
SimPhospho software tool
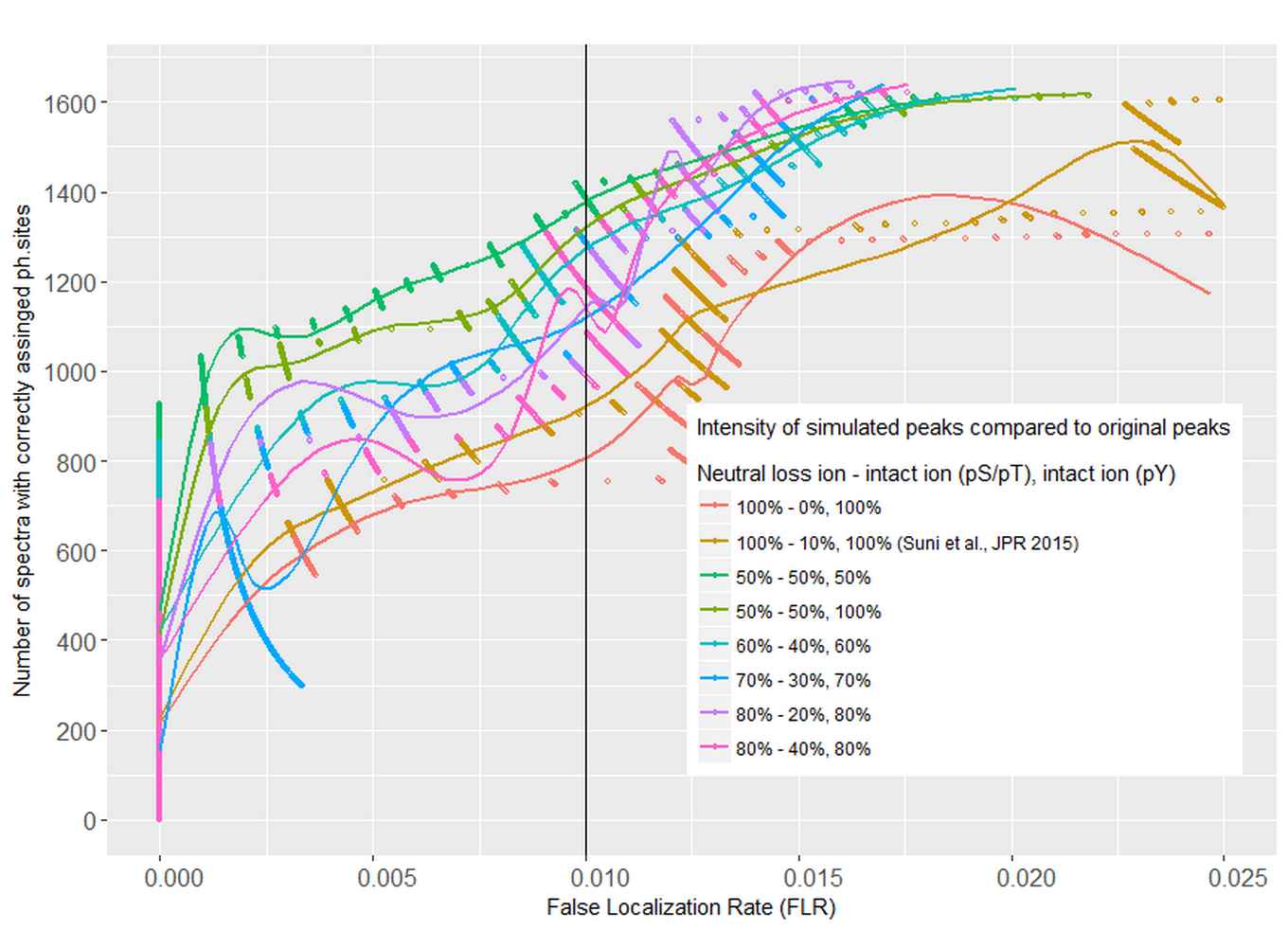
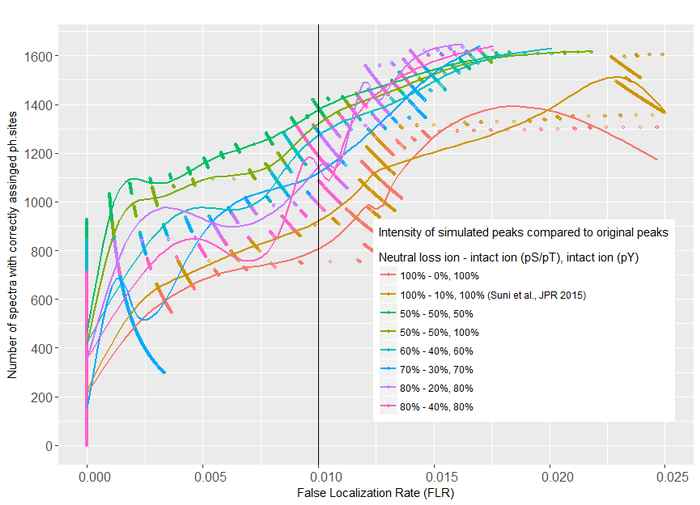
In an earlier study, the Dutch-Finnish team developed a method for tandem mass spectrometry interpretation based on the simulation of phosphopeptide spectral libraries, enabling highly sensitive and accurate phosphosite assignments. To promote more widespread use of this method, they now introduce SimPhospho, a fast and user-friendly tool for accurate simulation of phosphopeptide tandem mass spectra.
With SimPhospho, simulated phosphopeptide spectral libraries are used to validate and supplement database search results. It thus can improve reliable phosphoproteome identification and reporting. The program can be easily used together with the Trans-Proteomic Pipeline and it can be integrated in a phosphoproteomics data analysis workflow.
Open source
SimPhospho is open source and it is available for Windows, Linux and Mac operating systems. The software and its user’s manual with detailed description of data analysis as well as test data can be found at https://sourceforge.net/projects/simphospho/. A description of SimPhospho has just been published as an advance article in the journal Bioinformatics:
Veronika Suni, Tomi Suomi, Tomoya Tsubosaka, Susumu Y Imanishi, Laura L Elo, Garry L Corthals; SimPhospho: a software tool enabling confident phosphosite assignment, Bioinformatics, bty151, DOI: 10.1093/bioinformatics/bty151