Bernd Ensing receives eScience grant
1 February 2023
Designing new molecules is at the core of chemistry. In practice, however, creating molecular structures with useful properties is a difficult and time-consuming task. There are simply too many ways to connect different atoms to form new molecules - most of which are useless.
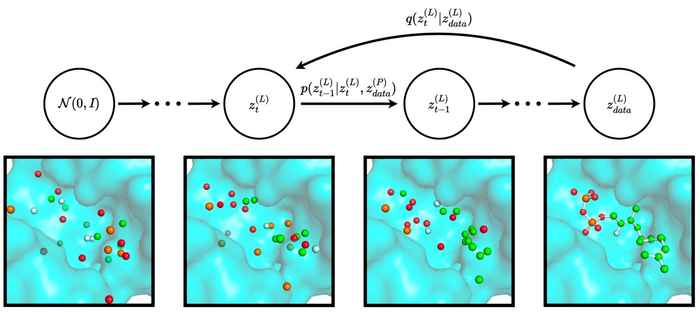
In the eScience project DeepMolGen, Ensing proposes to use a new class of machine learning techniques for generating new interesting molecules. These so-called diffusion models have already proven to be very useful in generating artistic images, movies, music, and speech. Recently the use of these models has also been reported for generating pharmaceutical drugs. Ensing and co-workers at the Computational Chemistry research group will develop and apply diffusion models in a variety of molecular use cases. They aim to recover the atomistic details from low-resolution simulations of polymers; optimize enzyme structures; and design better catalyst materials for the conversion of CO2 into useful chemicals. The eScience software engineer will assist in this development for a total of 0,5 fte in the course of the coming year.