Unravelling molecular self-organisation using a machine learning approach
Algorithm reveals temperature-dependent change in the nucleation mechanism of methane hydrate
25 April 2023
The emergence of a new crystal from a spontaneously formed nucleus or the folding of a protein are examples of rare molecular events that proceed rapidly after long waiting times in seeming stability. Due to their rarity as well as the complexity of the molecular systems involved, it is very difficult to obtain a thorough understanding of these processes - even when using computer models. Because it is impossible to predict when a rare event will occur, these models resort to calculating the dynamics of the molecular systems in series of tiny molecular steps. This can take up to a billion steps in a single simulation, but often this is not even sufficient given the timescale of many relevant molecular processes.
The paper in Nature Computational Science reports how the research team found a solution to this by combining computer simulations and artificial intelligence. They developed a machine learning algorithm that learns how to sample rare events. Using deep learning it builds mathematical models, based on transition path sampling, that help identify tell-tale signs for impending molecular transitions. The algorithm thus is able to ‘home in’ on the brief moments of the actual transitions, which prevents wasting computational resources waiting for such events to occur.
Thus, the algorithm can be used to study rare events occurring on previously inaccessible timescales. By autonomously initialising and analysing the modelling data, the algorithm reduces the amount of input required from researchers. Furthermore, by distilling the learned models into a human-accessible form, via so-called symbolic regression, the algorithm aids researchers in understanding and generalizing the findings to broad classes of systems.
Methane hydrate crystals
The machine-learning approach was tested, among others, in the nucleation of gas hydrate crystals. This is the field of expertise of Peter Bolhuis and Arjun, who obtained his PhD on transition path sampling of clathrate hydrate formation with Bolhuis in 2021. It was demonstrated that the algorithm was able to learn how temperature controls the way in which methane hydrates form. Such hydrates are ice-like solids that form at low temperature and high pressure from a liquid mixture of water and methane. In this phase transition, water molecules assemble spontaneously into an intricate crystal lattice with regularly spaced cages filled by methane. Despite commercial relevance in natural gas processing, the mechanism of gas-hydrate formation remains not completely understood.
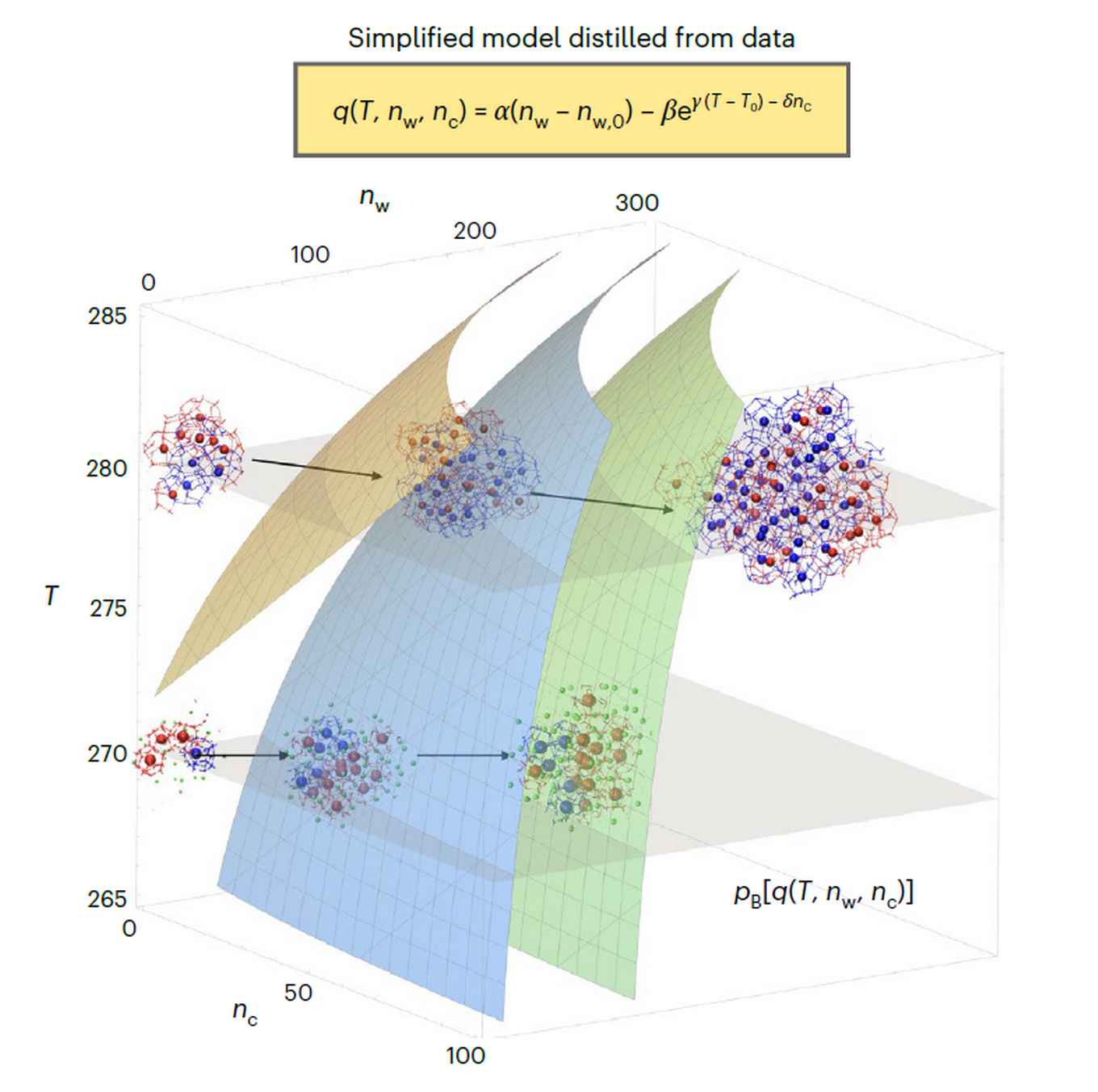
Using the novel machine learning approach, Bolhuis and Arjun were able to reveal a temperature-dependent change in the nucleation mechanism of methane hydrate. The data-driven, algorithm-generated mechanistic model predicted correctly how at low temperatures the size of the nucleus alone determines crystal growth. At higher temperatures, the number of a specific type of water cages gains in importance. Thus, a switch was established from amorphous growth at low temperatures to crystalline growth at higher temperatures.
The research was carried out in a cooperation between the Department of Theoretical Biophysics at the Max Planck Institute of Biophysics, the Institute for Advanced Studies (both in Frankfurt, Germany), the Institute of Biophysics at Goethe University Frankfurt, the Faculty of Physics at the University of Vienna (Austria) and the Van ’t Hoff Institute for Molecular Sciences at the University of Amsterdam (The Netherlands).
Abstract of the paper
Molecular self-organization driven by concerted many-body interactions produces the ordered structures that define both inanimate and living matter. Here we present an autonomous path sampling algorithm that integrates deep learning and transition path theory to discover the mechanism of molecular self-organization phenomena. The algorithm uses the outcome of newly initiated trajectories to construct, validate and—if needed—update quantitative mechanistic models. Closing the learning cycle, the models guide the sampling to enhance the sampling of rare assembly events. Symbolic regression condenses the learned mechanism into a human-interpretable form in terms of relevant physical observables. Applied to ion association in solution, gas-hydrate crystal formation, polymer folding and membrane-protein assembly, we capture the many-body solvent motions governing the assembly process, identify the variables of classical nucleation theory, uncover the folding mechanism at different levels of resolution and reveal competing assembly pathways. The mechanistic descriptions are transferable across thermodynamic states and chemical space.
Paper details
Hendrik Jung, Roberto Covino, A. Arjun, Christian Leitold, Christoph Dellago, Peter G. Bolhuis, Gerhard Hummer: Machine-guided path sampling to discover mechanisms of molecular self-organization. Nat. Comput. Sci. (2023). DOI: 10.1038/s43588-023-00428-z
See also
- Nature research briefing: A machine learning algorithm for studying how molecules self-assemble and function.
- Peter Bolhuis, research group Computational Chemistry.
- AI4Science at the University of Amsterdam.