Decoding the dissociation of sequence-specific protein–DNA complexes with non-equilibrium simulations
5 December 2023
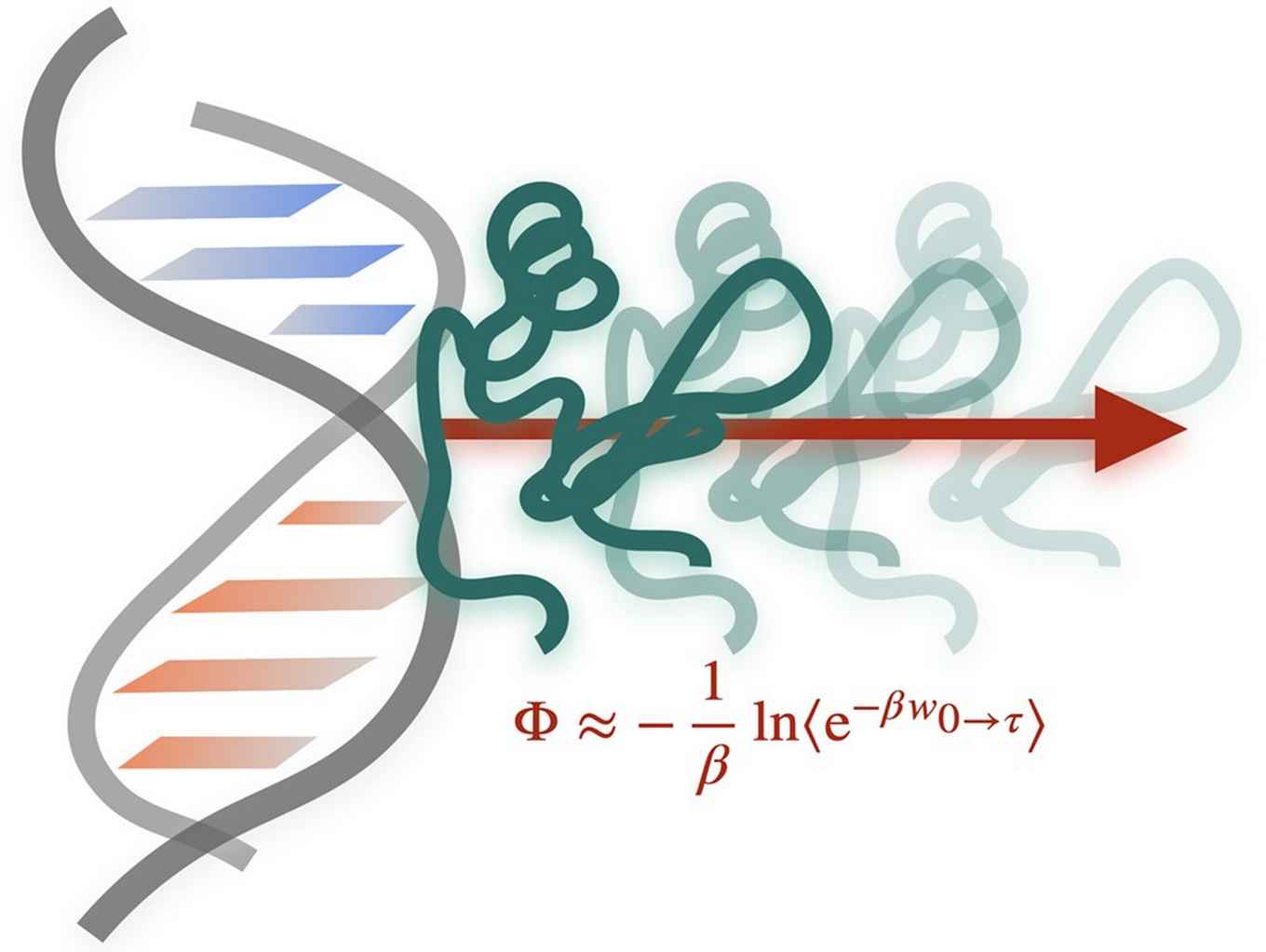
Sequence-specific protein–DNA interactions are crucial in processes such as DNA organization, gene regulation and DNA replication. Obtaining detailed insights into the recognition mechanisms of protein–DNA complexes through experiments is hampered by a lack of resolution in both space and time. In their paper in Nucleic Acids Research, the HIMS researchers present a molecular simulation approach to quantify the sequence specificity of protein–DNA complexes that yields results fast, and is generally applicable to any protein–DNA complex. The approach is based on molecular dynamics simulations in combination with a sophisticated steering potential and results in an estimate of the free energy difference of dissociation.
The simulations yield predictions about the affinity of DNA binding proteins for different nucleotide sequences, in agreement with experimental data. Specifically, the authors show this for the minor groove binding Histone-like Nucleoid Structuring (H-NS) protein and the major groove transcription factor ETS protein. Furthermore, the new approach offers mechanistic insights into the process of dissociation.
The protocol facilitates quantitative prediction of protein–DNA complex stability, while also providing high resolution insights into recognition mechanisms. As such, the simulation approach has the potential to yield detailed and quantitative insights into biological processes involving sequence-specific protein–DNA interactions.
Paper details
Thor van Heesch, Peter G Bolhuis, Jocelyne Vreede: Decoding dissociation of sequence-specific protein–DNA complexes with non-equilibrium simulations, Nucleic Acids Research, 2023, gkad1014. DOI: 10.1093/nar/gkad1014
See also
Research group Computational Chemistry