Unlocking novel therapies: cyclic peptide design for amyloidogenic targets
Advancement through synergy of experiments, simulations, and machine learning
16 January 2024
Current therapies for neurodegenerative diseases like Parkinson’s and Alzheimer’s disease address symptoms rather than preventing their onset. The work of Ilie and co-authors, supported by the Synapsis foundation, opens new doors for novel (preventive) therapeutic interventions and beyond. Particularly, it underlines the synergies between simulations, experiments, and machine learning when designing cyclic peptides as promising anti-amyloidogenic candidates. In the broader context, the proposed combination of strategies extends beyond cyclic peptide design, serving as a template for the de novo generation of any type of (bio)materials with programmable properties.
Abstract of the paper
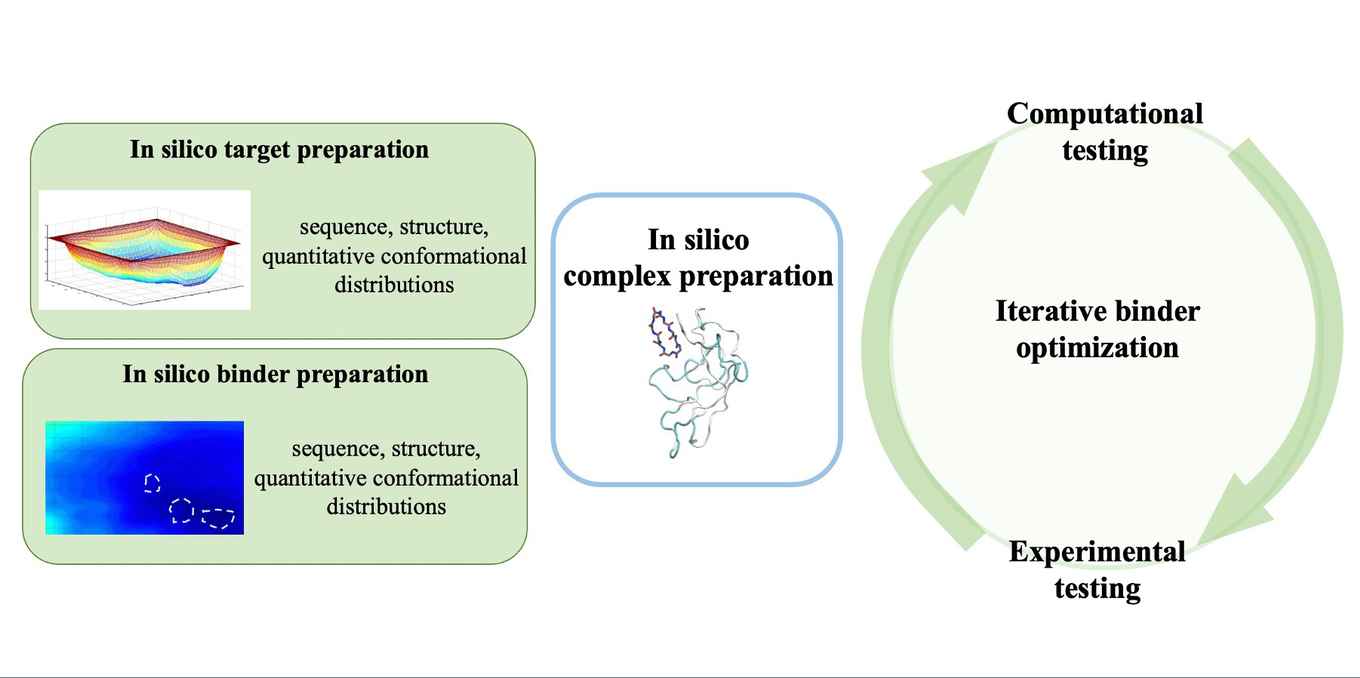
Existing therapies for neurodegenerative diseases like Parkinson's and Alzheimer's address only their symptoms and do not prevent disease onset. Common therapeutic agents, such as small molecules and antibodies struggle with insufficient selectivity, stability and bioavailability, leading to poor performance in clinical trials.
Peptide-based therapeutics are emerging as promising candidates, with successful applications for cardiovascular diseases and cancers due to their high bioavailability, good efficacy and specificity. In particular, cyclic peptides have a long in vivo stability, while maintaining a robust antibody-like binding affinity. However, the de novo design of cyclic peptides is challenging due to the lack of long-lived druggable pockets of the target polypeptide, absence of exhaustive conformational distributions of the target and/or the binder, unknown binding site, methodological limitations, associated constraints (failed trials, time, money) and the vast combinatorial sequence space. Hence, efficient alignment and cooperation between disciplines, and synergies between experiments and simulations complemented by popular techniques like machine-learning can significantly speed up the therapeutic cyclic-peptide development for neurodegenerative diseases.
We review the latest advancements in cyclic peptide design against amyloidogenic targets from a computational perspective in light of recent advancements and potential of machine learning to optimize the design process. We discuss the difficulties encountered when designing novel peptide-based inhibitors and we propose new strategies incorporating experiments, simulations and machine learning to design cyclic peptides to inhibit the toxic propagation of amyloidogenic polypeptides. Importantly, these strategies extend beyond the mere design of cyclic peptides and serve as template for the de novo generation of (bio)materials with programmable properties.
Paper details
Daria de Raffele & Ioana M. Ilie: Unlocking novel therapies: cyclic peptide design for amyloidogenic targets through synergies of experiments, simulations, and machine learning. Chem. Commun. 2024, 60, 632-645 DOI: 10.1039/D3CC04630C
The paper is part of the ChemCommun collection 2023 Emerging Investigators.
See also
Ilie research group: Multiscale simulations of biomolecular systems.